The following is a summary of my PhD thesis research, performed in the lab of Prof. Andre Levchenko, regarding the systems biology of information processing in the tumor necrosis factor (TNF) signaling network. A complete CV is available here.
-
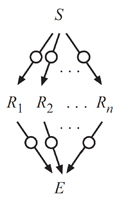
Information Theory
In collaboration with Dr. Ilya Nemenman (Emory), we pioneered the use of information theory to rigorously measure the limits that biochemical noise place on the fidelity of cell signaling. Based upon comprehensive single cell measurements, we determined that the TNF signaling network, as well as several other canonical signaling systems, could transmit up to just ~1 bit of information per use. The significance of this result is that it only allows a cell to accurately distinguish no more than two input values (e.g. presence or absence of TNF), thereby limiting the cell's ability to make nuanced decisions based upon signaling events. Furthermore, to explain the low capacity, we developed a general analytical framework based on information theory that correctly predicted the presence and precise magnitude of several previously unappreciated bottlenecks to information flow, such as receptor level noise and information loss due to negative feedback. Our results were published in Science and in PNAS.
-
Microfluidic analysis of signaling in single cells
Working with Dr. Joanne Wang, we developed a novel microfluidic device to enable high-throughput, high content analysis of individual cells, thereby allowing detailed examination of small numbers of genetically unperturbed cells exposed to multiple experimental conditions. The tool was featured on the cover of Science Signaling, and it allowed us to address a major controversy in the field, i.e. statistically ruling out asynchronous single cell oscillations as an underlying source for biphasic activity observed at the population level in TNF signaling. Due to the general utility of the technology, it also found widespread appeal in both academic and industry research. This allure led to several collaborations (e.g. examining epigenetic-specific signaling, and detecting bimodal JNK responses) as well as the formation of a startup to commercialize drug screening applications.
-
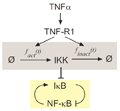
Predictive models of TNF-NF-κB signaling
I helped extend detailed ODE-based models of NF-κB responses to TNF and other inflammatory signals. These models generated high fidelity, experimentally verified predictions, thus providing a high degree of discovery potential and explanatory power. In collaboration with Dr. Alex Hoffmann (UCSD), I used models to reverse engineer a key molecular mechanism (rapid inhibition of the IκB kinase) regulating system dynamics in response to different doses of TNF, and demonstrated that such dynamics can enable spatially graded responses in a physiologically appropriate manner. In another project with Dr. Zhizhong Yin et al., I applied models to estimate the local amount of TNF delivered by a nanowire to an individual cell. These models have greatly influenced the field's attempts to quantitatively analyze TNF signaling.